Note: if you haven’t read Part 1 yet, do that first! Or don’t – who am I to tell you what to do?
On my last post on this subject I left the reader (and myself) with more questions than I did answers. Here, I’ll try to begin to resolve some of these questions without raising too many more (though I’ll likely fail at that).
As a brief recap: WLP300/WY3068, the strain that Weihenstephan uses for its hefeweizen (and has used since seemingly the beginning of time), is claimed by various sources to be one of two strains of yeast: S. cerevisiae or T. delbrueckii. There is definitely a dearth of information about this strain and whether it is one species or the other. There are a couple of scientific publications that claim that it is S. cerevisiae but don’t detail any in-depth species ID. The argument for WLP300 being T. delbrueckii is similarly weak, with all claims undocumented.
Via a cursory literature search, I found a couple of methods that might be useful in determining the species of this strain. One paper hinted that an osmotolerance experiment might be useful. However, upon a more thorough review it appears that the two strains of T. delbrueckii that were tested by this lab vary quite a bit in osmotolerance, to the point where this isn’t really a notable phenotype of the organism. So, that’s out the window – this is not a useful experiment to perform to determine the species of WLP300.
Another paper I found mentioned that T. delbrueckii will sporulate on normal nutrient rich media (YPD), while S. cerevisiae cannot (sporulation of both species can be induced using a starving the cells on a special media that is aptly named sporulation media). I decided to test this using a control strain of brewer’s yeast alongside some WLP300. I plated these cells out on YPD and let them sit for 15 days at 30C.
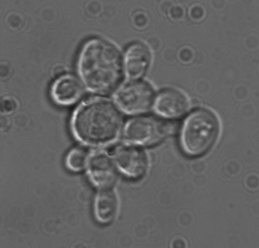
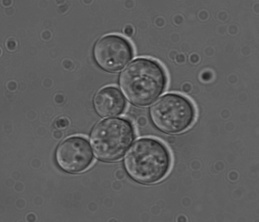
Unfortunately, I don’t have a T. delbrueckii strain on hand, although it would have been nice to have one to confirm the claim that this species can sporulate on YPD. At both 8 and 15 days, I pulled a small sample and checked them for the presence of sporulating yeast cells, which should look like panels A, C and D here. I’ve attached some photos of the WLP300 and a control strain on day 15 of growth on YPD (apologies in advance for the mediocre pictures taken with a dusty CCD).
I looked at several hundred cells for each sample and didn’t see a single sporulating cell, which is to be expected if both strains are S. cerevisiae. While these data certainly seem to indicate that WLP300 is a S. cerevisiae and not T. delbrueckii, it’s not quite enough information – it’s really just a part of the puzzle.
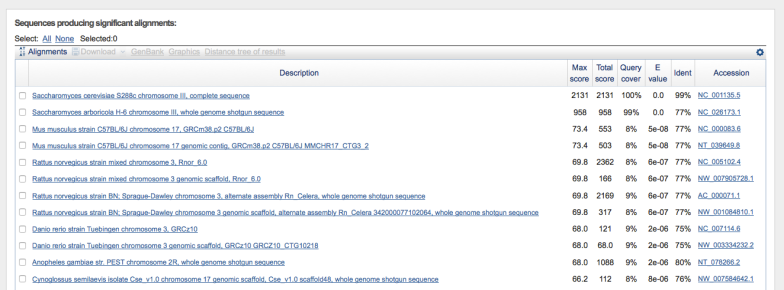
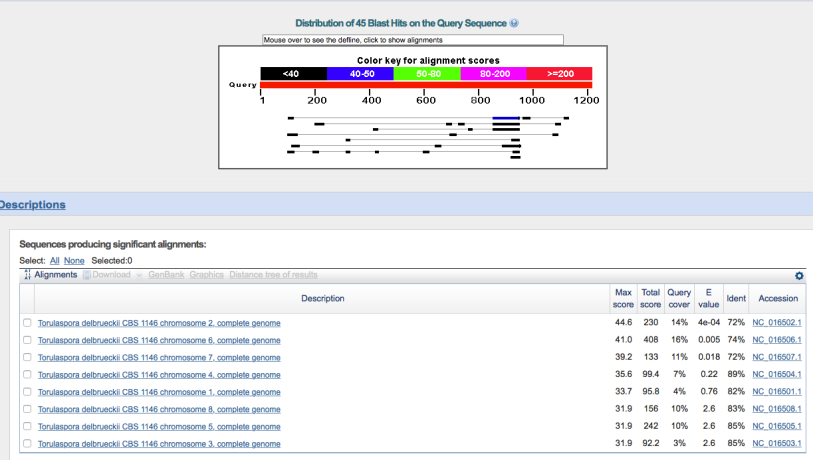
To further determine the likely species of WLP300, I pulled up some published sequencing data that was pointed out to me by reddit user thewhaleshark. There are 3 genes from the WLP300 genome that have freely accessible sequence data: RNQ1, YGL108C-like protein and Trp1. These data are easily accessible from this page. From there, it’s pretty straightforward to pull up a sequence from WLP300 in GenBank, and to BLAST it* against the entire collection of reference genomes (all species in the GenBank database).
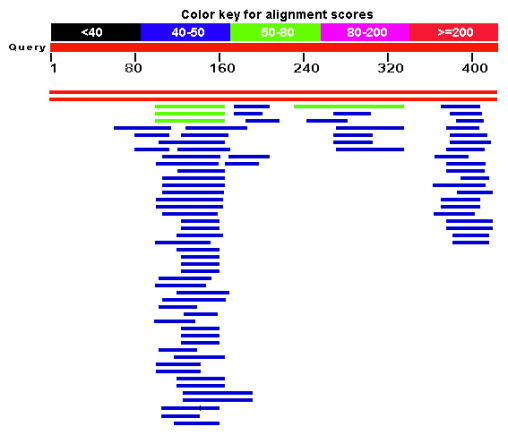
I pulled all 3 sequences, and BLASTed them against both genomes, using the BLASTn algorithm – this allows me to search for somewhat similar sequences instead of just perfect or very close matches. In other words, it reduces the stringency, allowing me to see if there is anything even semi-close to my query sequence. The results look something like this, an interactive list with the hits in order of rank. Red and longer bars are better hits, blue or black and shorter are worse:
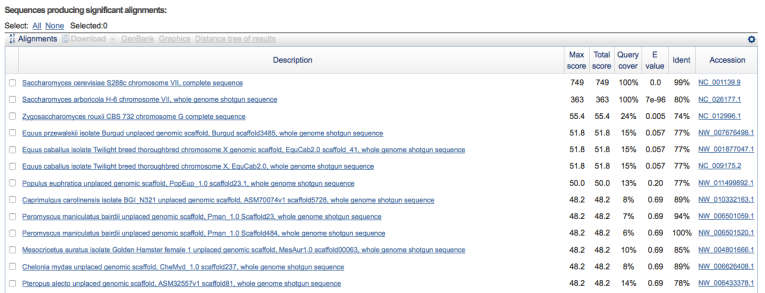
In addition, you get a list of the alignments that looks like this:
As you can see, S. cerevisiae ranks as the closest strain by a large margin, with the highest similarity score.
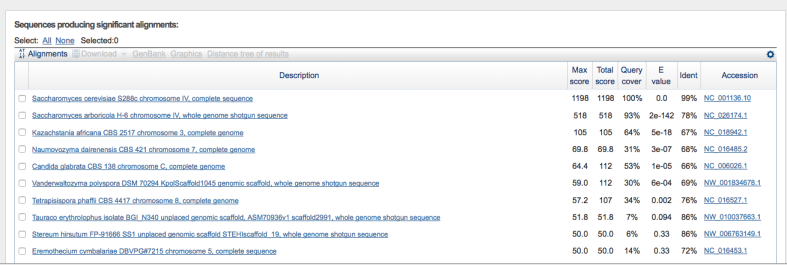
And here are the results for RNQ1 and Trp1, respectively:
So what can we conclude from this information? There is a very good chance that WLP300 is in fact S. cerevisiae and not T. delbrueckii. In fact, scrolling down the list, I didn’t observe any matches to the T. delbrueckii reference genome among the (very) distantly related hits.
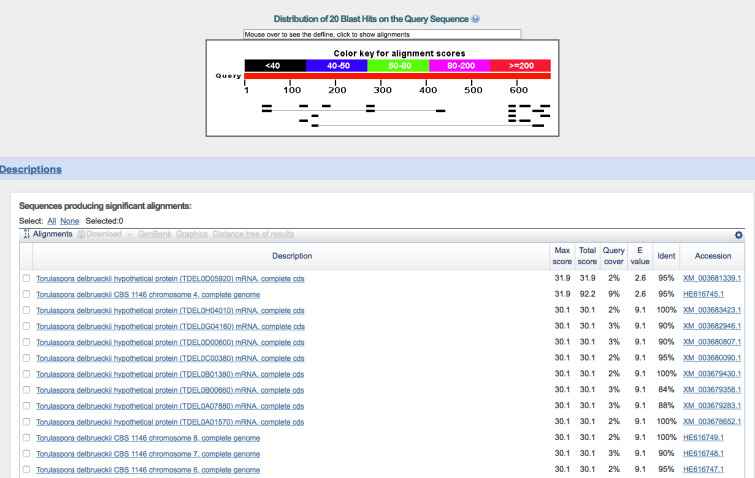
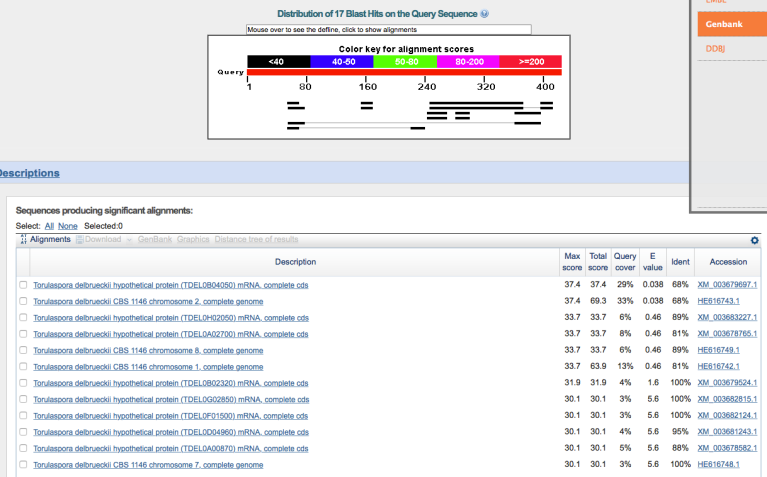
Out of curiosity, I wanted to determine how closely these genes aligned to anything in the T. delbrueckii genome, so I repeated this search but specified T. delbrueckii (taxid: 4950) as the only organism to query, forcing BLAST only to compare to T. delbrueckii.
So what do the above images indicate? These three genes in WLP300 are (by far) best aligned with the S. cerevisiae reference genome, and they do not align to any significant degree to the T. delbrueckii reference genome.¹ It’s not a nail in the coffin, but it is very hard to envision all three of these sequences aligning so poorly if WLP300 is indeed T. delbrueckii.
While these data were pretty convincing, I decided to contact some people who surely know more about WLP300 than I do in order to get their positions on the matter.
To check out one of the more knowledgable sources on this side of the pond, I got in touch with an rep at White Labs via email, who told me:
“We are in process of doing DNA sequencing on our strains. We will be publishing that when it is available. So far though, it appears to be S. cerevisiae.“
In a follow up email, they confirmed that this was based on previous characterization as well as the newer sequencing data.
At the urging of commenter Estus (in a comment on Part 1), I also contacted the Department of Brewing and Beverage Technology at TUM, research program associated with Weihenstephan, to ask them what species this strain is (known over there as St 68). They told me that St 68 is, without a doubt, S. cerevisiae.² People that have spent far more knowledge of this yeast strain than I have are convinced that it is S. cerevisiae, and I’m pretty sure that I agree with them based on the available data.
Another brief thought is that there are many people out there on the internet and in publications that claim that (in general, not necessarily WLP300 specifically) hefeweizen yeast are T. delbrueckii. Are any of these strains really T. delbrueckii? Perhaps the most notable claim that I can find is in Randy Mosher’s Tasting Beer. It also appears in a Google preview of his 2015 book “Mastering Homebrew,” although I am not sure if that is the final version of the book. I contacted him regarding his thoughts on this and he (graciously and rapidly) responded with the following:
That was based on the best available material I had at the time, and I would definitely defer to White Labs and other genuine experts in the area.
I have to say that yeast taxonomy is maddeningly contradictory and seems to change every decade or so. Hopefully gene-sequencing work like the research White Labs is helping with will clarify the situation once and for all.
Regarding that second to last point, I absolutely agree. It seems to be hard enough for taxonomists to keep up with microorganisms to ensure that they are appropriately categorized (especially given the capability to obtain increasingly detailed genomic datasets). It’s probably even harder for brewers to keep up with the taxonomists!
So where does this leave us? Well, if we broaden the scope a bit to all hefeweizen strains, I’m very interested in what differentiates hefeweizen strains from the rest of the family of ale yeast. It’s pretty obvious that they are genetically different to, say, WLP001, but I’d really love to see what those differences are. I’d hypothesize that the hefe yeast strains cluster together in a nice little subfamily on the yeast family tree. Who knows? Maybe they would cluster enough to justify their own species/subspecies. It’s impossible to know how genetically unique these yeast strains really are without whole-genome data. However, the requisite data for elucidating these finer points will (or already does) exist courtesy of White Labs.
Ultimately, does it really matter what species WLP300 is? To most people, probably not. It makes a great beer, whether it is S. cerevisiae or T. delbrueckii. However, we’re always learning new things about the various yeast strains that we brew with, and I personally find it pretty entertaining to ask these kinds of questions. I’ll certainly be keeping an eye out for the data coming from White Labs/illumina/Synthetic Genomics – the data that come from that project should be very, very interesting.
* For those less familiar with genetics, BLAST stands for “Basic Local Alignment Search Tool.” It allows scientists to look for sequences in a standardized genome (in this case, from S. cerevisiae or T. delbrueckii). It is essentially a genomic search engine. It allows you to match up your query sequence with matching sequences in a genome, and ranks the results based on similarity to your query sequence.
¹ Even though it doesn’t appear that WLP300 is a T.delbrueckii, it surprised me that it was so different – I was expecting there to be a little bit more homology considering these two species were originally classified within the Saccharomyces genus.
² As a side note, the contact at TUM said that have encountered T. delbrueckii as an unwanted strain in beer making but also mentioned that it does have practical/volitional applications in wineries.
Thanks for sharing your information. Nice work. Which all might support my initial idea that WY3068 was unintentionally listed as T. delbrueckii rather than S. delbrueckii (synonym of S. cerevisiae). I used MrMalty as my reference for the taxonomy and interesting enough, in MrMalty’s list WY3068 is now listed as S. delbrueckii. Futhermore, I would not be surprised if it somehow turns out that T. delbrueckii is not even linked to wheat beers. I for myself could not find any publication backing up this relationship.
Concerning the YPD and T. delbrueckii sporulation test. I have a sample of T. delbrueckii in my current stock as well as YPD media. Will plate some T. delbrueckii on YPD in case I do other YPD platings in the future and let you know what I came up with. Furthermore use T. delbrueckii in a fermentation test against other non-Saccharomyces yeasts. Keep on posting. Enjoy reading your blog.
LikeLike
Thanks – I really appreciate it. Regarding S. delbrueckii, I was under the impression that it was recategorized to T. delbrueckii and is not synonymous with S.cerevisae but I could be misinformed (http://www.cbs.knaw.nl/Collections/BioloMICS.aspx?Link=T&TableKey=14682616000000067&Rec=106649&Fields=All).
I agree with you that broadly, T. delbrueckii is likely not linked to wheat beers.
I’m very interested to see if it can sporulate on YPD – as I have no T. delbrueckii I cannot test that – I’ll be interested to see your results regarding that!
I, too, am planning on trying a T. delbrueckii based fermentation. Based on it’s use in the wine industry I suspect that it cannot generate enough alcohol alone to kill off bad bugs – it may need to be a mixed culture with S. cerevisae. As I’m sure you’ve seen, there are several publications regarding the used of mixed T. delbrueckii/S. cerevisae cultures in wine.
Thanks – I appreciate your thoughts!
LikeLike
> Regarding S. delbrueckii, I was under the impression that it was recategorized to T. delbrueckii and is not synonymous with S.cerevisae but I could be misinformed
Sorry, I must have mixed something up here. You are right. S. delbrueckii is a synonym for T. delbrueckii. Checked my Kurtzman. So my hypothesis seems to have a basic fault…
> I, too, am planning on trying a T. delbrueckii based fermentation. Based on it’s use in the wine industry I suspect that it cannot generate enough alcohol alone to kill off bad bugs – it may need to be a mixed culture with S. cerevisae. As I’m sure you’ve seen, there are several publications regarding the used of mixed T. delbrueckii/S. cerevisae cultures in wine.
Cool. Looking forward to read your results then. I will try my Torula in a mixed fermentation. Mainly the reason why I looked at the sugar composition of wort & posted about it. I previously tried another non-Sacc yeast in a mixed fermentation (against a Sacc control) and will post these results soon. I got my Torula from CHR Hansen (PRELUDE) http://www.chr-hansen.com/products/product-areas/wine-ingredients/viniflorar-specialty-yeasts-to-get-closer-to-wild-ferment-complexity-without-the-risk.html.
> Thanks – I appreciate your thoughts!
So do I. Keep up the good work. Always like to read about yeasts & science 🙂
LikeLiked by 1 person
Finally caught up on reading these two posts, good science and I am proceeding with my experiment to make a 100% T. delbrueckii beer with the CONFIRMED T. delbrueckii culture I obtained.
LikeLiked by 1 person
excellent – looking forward to hearing how it comes out. Hopefully it can tolerate a not-mixed culture!
life has been a bit hectic recently so I have not had the time to tend to this blog recently but I’m also hoping to put together a 100% t. delbrueckii beer in the near future; will definitely post about that when I do.
LikeLiked by 1 person
I am finishing up a 100% Kluyveromyces lactis beer (should be done in a few weeks I think) but I have obtained pure cultures with good providence of Hanseniaspora uvarum, Torulaspora delbrueckii, Pichia membranifaciens, Metschnikowia pulcherrima, and Zygosaccharomyces bailii. I’m going to do 100% fermentations of a simple wort with all these guys.
Since the attenuation of each of these species is likely going to be garbage, I am considering letting them ferment as much as they can (give them 6-8 weeks) and then pitch some neutral Sacc on top of it just to ferment out. That way they end up at a lower gravity but the “weird” organism had a chance to impart the flavor.
i totally understand about not keeping up with the blog. I didn’t realize it until a few days ago but apparently I took March off.
LikeLike
Sounds like a great project! Are you concerned about spoilage or pathogens in such a slow ferment?
LikeLike
It is a mild concern with anything but I can monitor it and just use good technique and hope for the best.
LikeLiked by 1 person
I figured as much; but if you don’t try, who will? Looking forward to reading about the results!
LikeLike